Perturbations of BMP/TGF-β and VEGF/VEGFR signalling pathways in non-syndromic sporadic brain arteriovenous malformations (BAVM)
- 新闻
Brain arteriovenous malformations (BAVM) represent a congenital anomaly of the cerebral vessels with a prevalence of 10–18/100 000. BAVM is the leading aetiology of intracranial haemorrhage in children. Our objective was to identify gene variants potentially contributing to disease and to better define the molecular aetiology underlying non-syndromic sporadic BAVM. We performed whole-exome trio sequencing of 100 unrelated families with a clinically uniform BAVM phenotype. Pathogenic variants were then studied in vivo using a transgenic zebrafish model. We identified four pathogenic heterozygous variants in four patients, including one in the established BAVM-related gene, ENG, and three damaging variants in novel candidate genes: PITPNM3, SARS and LEMD3, which we then functionally validated in zebrafish. In addition, eight likely pathogenic heterozygous variants (TIMP3, SCUBE2, MAP4K4, CDH2, IL17RD, PREX2, ZFYVE16 and EGFR) were identified in eight patients, and 16 patients carried one or more variants of uncertain significance. Potential oligogenic inheritance (MAP4K4 with ENG, RASA1 with TIMP3 and SCUBE2 with ENG) was identified in three patients. Regulation of sma- and mad-related proteins (SMADs) (involved in bone morphogenic protein (BMP)/transforming growth factor beta (TGF-β) signalling) and vascular endothelial growth factor (VEGF)/vascular endotheliual growth factor recepter 2 (VEGFR2) binding and activity (affecting the VEGF signalling pathway) were the most significantly affected biological process involved in the pathogenesis of BAVM. Our study highlights the specific role of BMP/TGF-β and VEGF/VEGFR signalling in the aetiology of BAVM and the efficiency of intensive parallel sequencing in the challenging context of genetically heterogeneous paradigm.
 Figure 1 Workflow of patient enrolment and analysis of whole-exome sequencing data
Figure 1 Workflow of patient enrolment and analysis of whole-exome sequencing data
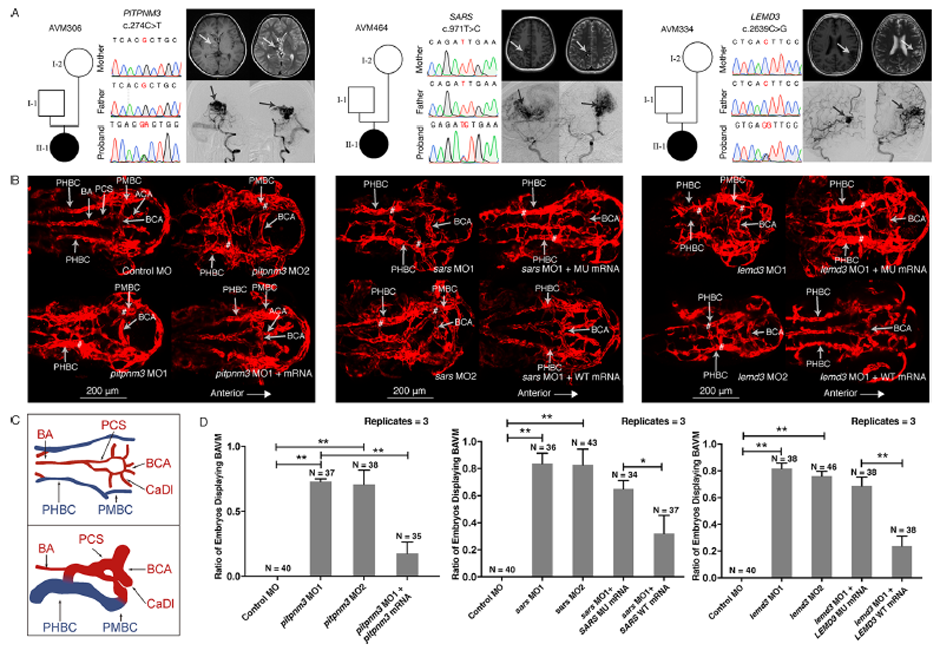 Figure 2 Variant information, phenotype and in vivo functional study of PITPNM3, SARS and LEMD3 variants in patient AVM306, AVM464 and AVM334.
Figure 2 Variant information, phenotype and in vivo functional study of PITPNM3, SARS and LEMD3 variants in patient AVM306, AVM464 and AVM334.
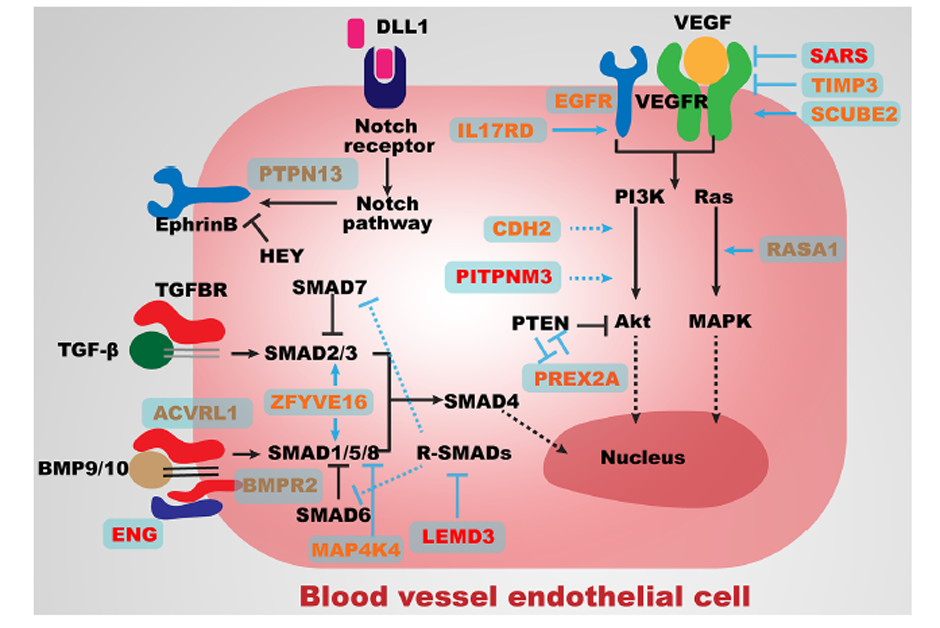 Figure 3 Major angiogenesis pathways associated with genes identified in the present study.
Figure 3 Major angiogenesis pathways associated with genes identified in the present study.

